Lomas: Client demo
This notebook showcases how researcher could use the Lomas platform. It explains the different functionnalities provided by the lomas-client library to interact with the secure server.
The secure data are never visible by researchers. They can only access to differentially private responses via queries to the server.
Each user has access to one or multiple projects and for each dataset has a limited budget with \(\epsilon\) and \(\delta\) values.
[ ]:
from IPython.display import Image
Image(filename="images/image_demo_client.png", width=800)
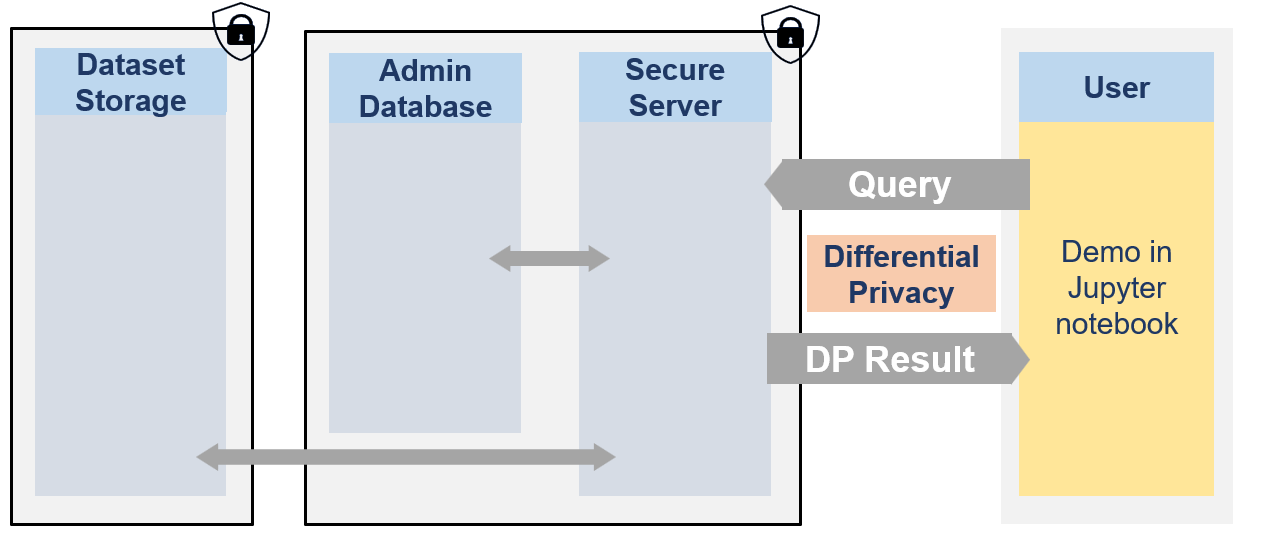
🐧🐧🐧 In this notebook the researcher is a penguin researcher named Dr. Antarctica. She aims to do a grounbdbreaking research on various penguins dimensions.
Therefore, the powerful queen Icerbegina 👑 had the data collected. But in order to get the penguins to agree to participate she promised them that no one would be able to look at the data and that no one would be able to guess the bill width of any specific penguin (which is very sensitive information) from the data. Nobody! Not even the researchers. The queen hence stored the data on the Secure Data Disclosure Server and only gave a small budget to Dr. Antarctica.
This is not a problem for Dr. Antarctica as she does not need to see the data to make statistics thanks to the Secure Data Disclosure Client library lomas-client. 🐧🐧🐧
Step 1: Install the library
To interact with the secure server on which the data is stored, Dr.Antartica first needs to install the library lomas-client on her local developping environment.
It can be installed via the pip command:
[ ]:
# !pip install lomas-client
[ ]:
%load_ext autoreload
%autoreload 2
from lomas_client import Client
import numpy as np
/home/aymond/work/lomas/lomas/.devenv/state/venv/lib/python3.13/site-packages/opendp_logger/deserialize.py:7: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
import pkg_resources
Step 2: Initialise the client
Once the library is installed, a Client object must be created. It is responsible for sending sending requests to the server and processing responses in the local environment. It enables a seamless interaction with the server.
The client needs a few parameters to be created. Usually, these would be set in the environment by the system administrator (queen Icebergina) and be transparent to lomas users. In this instance, the following code snippet sets a few of these parameters that are specific to this notebook.
She will only be able to query on the real dataset if the queen Icebergina has previously made her an account in the database, given her access to the PENGUIN dataset and has given her some epsilon and delta credit.
[ ]:
# The following would usually be set in the environment by a system administrator
# and be tranparent to lomas users.
# Uncomment them if you are running against a Kubernetes deployment.
# They have already been set for you if you are running locally within a devenv or the Jupyter lab set up by Docker compose.
import os
# os.environ["LOMAS_CLIENT_APP_URL"] = "https://lomas.example.com:443"
# os.environ["LOMAS_CLIENT_OIDC_DISCOVERY_URL"] = "https://dex.example.com:443/.well-known/openid-configuration"
# os.environ["LOMAS_CLIENT_TELEMETRY__ENABLED"] = "false"
# os.environ["LOMAS_CLIENT_TELEMETRY__COLLECTOR_ENDPOINT"] = "http://otel.example.com:445"
# os.environ["LOMAS_CLIENT_TELEMETRY__COLLECTOR_INSECURE"] = "true"
# os.environ["LOMAS_CLIENT_TELEMETRY__SERVICE_ID"] = "my-app-client"
# os.environ["LOMAS_CLIENT_REALM"] = "lomas"
# We set these ones because they are specific to this notebook.
os.environ["LOMAS_CLIENT_USER_NAME"] = "dr.antartica@example.com"
os.environ["LOMAS_CLIENT_USER_PASSWORD"] = "dr.antartica"
os.environ["LOMAS_CLIENT_DATASET_NAME"] = "PENGUIN"
# Note that all client settings can also be passed as keyword arguments to the Client constructor.
# eg. client = Client(user_name = "Dr.Antartica") takes precedence over setting the "LOMAS_CLIENT_USER_NAME"
# environment variable.
[ ]:
client = Client()
[15:28:02] WARNING Keycloak or Lomas service configured without TLS -> using oauthlib insecure http_client.py:31 transport
And that’s it for the preparation. She is now ready to use the various functionnalities offered by lomas_client.
Step 3: Understand the functionnalities of the library
a. Getting dataset metadata
Dr. Antartica has never seen the data and as a first step to understand what is available to her, she would like to check the metadata of the dataset. Therefore, she just needs to call the get_dataset_metadata() function of the client. As this is public information, this does not cost any budget.
This function returns metadata information in a format based on SmartnoiseSQL dictionary format, where among other, there is information about all the available columns, their type, bound values (see Smartnoise page for more details). Any metadata is required for Smartnoise-SQL is also required here and additional information such that the different categories in a string type column column can be added.
[ ]:
penguin_metadata = client.get_dataset_metadata()
penguin_metadata
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_dataset_metadata' with query params: {'dataset_name': 'PENGUIN'}.
{'max_ids': 1,
'rows': 344,
'row_privacy': True,
'censor_dims': False,
'clamp_counts': True,
'clamp_columns': True,
'use_dpsu': False,
'columns': {'species': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 3,
'categories': ['Adelie', 'Chinstrap', 'Gentoo']},
'island': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 3,
'categories': ['Torgersen', 'Biscoe', 'Dream']},
'bill_length_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 30.0,
'upper': 65.0,
'int_with_nulls': False},
'bill_depth_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 13.0,
'upper': 23.0,
'int_with_nulls': False},
'flipper_length_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 150.0,
'upper': 250.0,
'int_with_nulls': False},
'body_mass_g': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 2000.0,
'upper': 7000.0,
'int_with_nulls': True},
'sex': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 2,
'categories': ['MALE', 'FEMALE']}}}
Based on this Dr. Antartica knows that there are 7 columns, 3 of string type (species, island, sex) with their associated categories (i.e. the species column has 3 possibilities: ‘Adelie’, ‘Chinstrap’, ‘Gentoo’) and 4 of float type (bill length, bill depth, flipper length and body mass) with their associated bounds (i.e. the body mass of penguin ranges from 2000 to 7000 gramms). She also knows based on the field max_ids: 1 that each penguin can only be once in the dataset and on the field
row_privacy: True that each row represents a single penguin. Finally, she learns that there are 344 rows in the dataset and hence 344 penguins.
[ ]:
NB_PENGUINS = penguin_metadata["rows"]
b. Get a dummy dataset
Now, that she has seen and understood the metadata, she wants to get an even better understanding of the dataset (but is still not able to see it). A solution to have an idea of what the dataset looks like it to create a dummy dataset.
Based on the public metadata of the dataset, a random dataframe can be created created. By default, there will be 100 rows and the seed is set to 42 to ensure reproducibility, but these 2 variables can be changed to obtain different dummy datasets. Getting a dummy dataset does not affect the budget as there is no differential privacy here. It is not a synthetic dataset and all that could be learn here is already present in the public metadata (it is created randomly on the fly based on the metadata).
Dr. Antartica first create a dummy dataset with 100 rows and chooses a seed of 0.
[ ]:
NB_ROWS = 100
SEED = 0
[ ]:
df_dummy = client.get_dummy_dataset(
nb_rows = NB_ROWS,
seed = SEED
)
print(df_dummy.shape)
df_dummy.head()
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_dummy_dataset' with query params: {'dataset_name': 'PENGUIN', 'dummy_nb_rows': 100, 'dummy_seed': 0}.
(100, 7)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Gentoo | Biscoe | 46.799577 | 16.196816 | 239.680123 | 3011.0 | FEMALE |
| 1 | Chinstrap | Dream | 38.133052 | 14.875077 | 208.332005 | 6690.0 | MALE |
| 2 | Chinstrap | Torgersen | 58.065820 | 19.725266 | 154.021822 | 2474.0 | MALE |
| 3 | Adelie | Torgersen | 62.323556 | 14.951074 | 221.148682 | 2024.0 | FEMALE |
| 4 | Adelie | Dream | 39.314560 | 18.776879 | 206.902585 | 3615.0 | MALE |
c. Check privacy loss budget ε, δ (initial, current, remaining)
It is the first time that Dr. Antartica connects to the server and she wants to know how much buget has beeen assigned to her. Therefore, she calls the fonction get_initial_budget.
[ ]:
client.get_initial_budget()
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_initial_budget' with query params: {'dataset_name': 'PENGUIN'}.
InitialBudgetResponse(initial_epsilon=10.0, initial_delta=0.005)
She sees that she has 10.0 epsilon and 0.005 epsilon at her disposal.
Then she checks her total spent budget get_total_spent_budget. As she only did queries on metadata on dummy dataframes, this should still be 0.
[ ]:
client.get_total_spent_budget()
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_total_spent_budget' with query params: {'dataset_name': 'PENGUIN'}.
SpentBudgetResponse(total_spent_epsilon=0.0, total_spent_delta=0.0)
It will also be useful to know what the remaining budget is. Therefore, she calls the function get_remaining_budget. It just substarcts the total spent budget from the initial budget.
[ ]:
client.get_remaining_budget()
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_remaining_budget' with query params: {'dataset_name': 'PENGUIN'}.
RemainingBudgetResponse(remaining_epsilon=10.0, remaining_delta=0.005)
As expected, for now the remaining budget is equal to the inital budget.
Step 4: Use DP libraries to analyse the dataset
Available DP libraires are:
Smartnoise-SQL for SQL-like queries
Smartnoise-Synth for generating synthetic datasets
OpenDP for summary statistics
DiffPrivLib for training Machine Learning models
For each library, there are three possibilities:
estimate the cost of a query (will NOT spend privacy loss budget)
query on a ‘dummy’ dataset (explained below) (will NOT spend privacy loss budget)
query on the private dataset (WILL SPEND PRIVACY LOSS BUDGET)
a. Compute average bill length with Smartnoise-SQL
Dr. Antartica wants to know the average bill length of penguins. Therefore, she will use smartnoise-sql library and write the associated SQL command.
[ ]:
# Average bill length in mm
QUERY = "SELECT AVG(bill_length_mm) AS avg_bill_length_mm FROM df"
Estimate cost of a query with smartnoise-sql
She will then estimate the cost of this query. In the various DP librairies the budget that will by used by a query in the server might be slightly different than what is asked by the user in inptu. The estimate cost function of each library returns the cost that will effectively be sent and deduced if the query is applied on the sensitive dataset.
The user can then decide to use the budget or modify it. Again, of course, this will not impact the user’s budget.
Dr. Antartica checks the budget that computing the average bill length will really cost her if she asks the query with an epsilon and a delta.
[ ]:
EPSILON = 0.5
DELTA = 1e-4
[ ]:
cost = client.smartnoise_sql.cost(
query = QUERY,
epsilon = EPSILON,
delta = DELTA
)
cost
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'estimate_smartnoise_sql_cost' with query params: {'dataset_name': 'PENGUIN', 'query_str': 'SELECT AVG(bill_length_mm) AS avg_bill_length_mm FROM df', 'epsilon': 0.5, 'delta': 0.0001, 'mechanisms': {}}.
CostResponse(epsilon=1.0, delta=4.999999999999449e-05)
[ ]:
print(f"This query would actually cost her {cost.epsilon} epsilon and {cost.delta} delta.")
This query would actually cost her 1.0 epsilon and 4.999999999999449e-05 delta.
She decides that it is good enough.
Query average bill length on dummy dataset with smartnoise-sql
She now wants to start querying the real dataset for her research.
However, her budget is limited and it would be a waste to spend it by mistake on a coding error. Therefore the client/server pipeline has functionnal testing capabilities for the users. It is possible to test a query on a dummy dataset to ensure that everything is working properly. Dr. Antartica will not be able to use the results of a dummy query for her analysis (as the data is random) but if the query on the dummy dataset works, she can be confident that her query will also work on the
real dataset. This functionnal testing on the dummy does not have any impact on the budget as it is on random data only.
To test on the dummy data instead of the real data, the function call is exactly the same with the only exception of the flag dummy=True. In the following cell, she will test with smartnoise_query but it is the same flag for opendp.query. She can optionnaly give two additional parameters to set the seed and the number of rows of the dummy dataset.
Another more advanced possibility for functionnal tests with the dummy is to compare results of queries on a local dummy and the remote dummy with a very high budget:
create a local dummy on the notebook with a specific seed and number of rows
compute locally the wanted query on this local dummy with python functions like numpy
query the server on the same remote dummy with (
dummy=True, same seed and same number of row) and a very big buget to limit noise as much as possible (don’t worry this won’t cost any real budget)compare and verify that the local and remote dummy have similar results.
Dr. Antartica will follow the best practice and now try the query to get the average bill length (in mm) on the dummy dataset. She does not forget to
set the
dummyflag to Trueset very high budget values to be able to compare results with a similar local dummy (with the same seed and number of rows) if she wants to verify that the function do what is expected. Here she will just check that the number of rows is close to what she sets as parameter.
[ ]:
# On the remote server dummy dataframe
dummy_res = client.smartnoise_sql.query(
query = QUERY,
epsilon = 100.0,
delta = 0.99,
dummy = True,
nb_rows = NB_ROWS,
seed = SEED
)
[15:28:04] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'dummy_smartnoise_sql_query' with query params: {'dataset_name': 'PENGUIN', 'dummy_nb_rows': 100, 'dummy_seed': 0, 'query_str': 'SELECT AVG(bill_length_mm) AS avg_bill_length_mm FROM df', 'epsilon': 100.0, 'delta': 0.99, 'mechanisms': {}, 'postprocess': True}.
[ ]:
print(f"Average bill length in remote dummy: {np.round(dummy_res.result.df['avg_bill_length_mm'][0], 2)}mm.")
Average bill length in remote dummy: 48.58mm.
No functionnal errors happened and the average bill length is within reasonable bounds. She is now even more confident in using her query on the server.
Query average bill length on private dataset with smartnoise-sql
Now that all the safeguard functions were tested, Dr. Antartica is ready to query on the real dataset and get a differentially private response of the number of penguins and average bill length. By default, the flag dummy is False so setting it is optional. She uses the values of epsilon and delta that she selected just before.
Careful: This command DOES spend the budget of the user and the remaining budget is updated for every query.
[ ]:
client.get_remaining_budget()
[15:28:06] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_remaining_budget' with query params: {'dataset_name': 'PENGUIN'}.
RemainingBudgetResponse(remaining_epsilon=10.0, remaining_delta=0.005)
[ ]:
response = client.smartnoise_sql.query(
query = QUERY,
epsilon = EPSILON,
delta = DELTA,
dummy = False # APPLIED ON SENSITIVE DATA, WILL SPEND BUDGET
)
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'smartnoise_sql_query' with query params: {'dataset_name': 'PENGUIN', 'query_str': 'SELECT AVG(bill_length_mm) AS avg_bill_length_mm FROM df', 'epsilon': 0.5, 'delta': 0.0001, 'mechanisms': {}, 'postprocess': True}.
[ ]:
avg_bill_length = np.round(response.result.df['avg_bill_length_mm'].iloc[0], 2)
print(f"Average bill length of penguins in real data: {avg_bill_length}mm.")
Average bill length of penguins in real data: 44.29mm.
After each query on the real dataset, the budget informations are also returned to the researcher. It is possible possible to check the remaining budget again afterwards:
[ ]:
client.get_remaining_budget()
[15:28:07] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_remaining_budget' with query params: {'dataset_name': 'PENGUIN'}.
RemainingBudgetResponse(remaining_epsilon=9.0, remaining_delta=0.004950000000000006)
As can be seen in get_total_spent_budget(), it is the budget estimated with estimate_smartnoise_cost() that was spent.
[ ]:
client.get_total_spent_budget()
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_total_spent_budget' with query params: {'dataset_name': 'PENGUIN'}.
SpentBudgetResponse(total_spent_epsilon=1.0, total_spent_delta=4.999999999999449e-05)
Dr. Antartica has now a differentially private estimation of the number of penguins in the dataset and is confident to use the library for the rest of her analyses.
b. Compute confidence interval with opendp
[ ]:
import opendp as dp
import opendp.transformations as trans
import opendp.measurements as meas
She now wants the confidence interval of bill length in mm. She already has the number of penguins and the average from the metadata and previous smartnoise-sql queries respectively. She now needs the variance value.
Prepare opendp pipeline and verify on dummy
She checks the metadata of the columns again to use the relevant values in the pipeline.
[ ]:
penguin_metadata["columns"]
{'species': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 3,
'categories': ['Adelie', 'Chinstrap', 'Gentoo']},
'island': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 3,
'categories': ['Torgersen', 'Biscoe', 'Dream']},
'bill_length_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 30.0,
'upper': 65.0,
'int_with_nulls': False},
'bill_depth_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 13.0,
'upper': 23.0,
'int_with_nulls': False},
'flipper_length_mm': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 150.0,
'upper': 250.0,
'int_with_nulls': False},
'body_mass_g': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'float',
'precision': 64,
'lower': 2000.0,
'upper': 7000.0,
'int_with_nulls': True},
'sex': {'private_id': False,
'nullable_proportion': 0.0,
'max_partition_length': None,
'max_influenced_partitions': None,
'max_partition_contributions': None,
'type': 'string',
'cardinality': 2,
'categories': ['MALE', 'FEMALE']}}
She can define the columns names and the bounds of the relevant column.
[ ]:
columns = list(penguin_metadata["columns"].keys())
[ ]:
bill_length_min = penguin_metadata['columns']['bill_length_mm']['lower']
bill_length_max = penguin_metadata['columns']['bill_length_mm']['upper']
bill_length_min, bill_length_max
(30.0, 65.0)
She can now define the pipeline of the transformation to have the variance that she wants on the data:
[ ]:
bill_length_transformation_pipeline = (
trans.make_split_dataframe(separator=",", col_names=columns) >>
trans.make_select_column(key="bill_length_mm", TOA=str) >>
trans.then_cast_default(TOA=float) >>
trans.then_clamp(bounds=(bill_length_min, bill_length_max)) >>
trans.then_resize(size=NB_PENGUINS, constant=avg_bill_length) >>
trans.then_variance()
)
However, when she tries to execute it on the server, she has an error (see below).
[ ]:
# No instruction for noise addition mechanism: Expect to fail !!!
client.opendp.query(
opendp_pipeline = bill_length_transformation_pipeline,
dummy=True
)
---------------------------------------------------------------------------
InvalidQueryException Traceback (most recent call last)
Cell In[29], line 2
1 # No instruction for noise addition mechanism: Expect to fail !!!
----> 2 client.opendp.query(
3 opendp_pipeline = bill_length_transformation_pipeline,
4 dummy=True
5 )
File ~/work/lomas/lomas/client/lomas_client/libraries/opendp.py:139, in OpenDPClient.query(self, opendp_pipeline, fixed_delta, mechanism, dummy, nb_rows, seed)
103 def query(
104 self,
105 opendp_pipeline: dp.Measurement | pl.LazyFrame,
(...) 110 seed: int = DUMMY_SEED,
111 ) -> QueryResponse:
112 """This function executes an OpenDP query.
113
114 Args:
(...) 137 QueryResponse: A dictionary of the response body containing the deserialized pipeline result.
138 """
--> 139 body_json = self._get_opendp_request_body(
140 opendp_pipeline,
141 fixed_delta=fixed_delta,
142 mechanism=mechanism,
143 )
145 request_model: type[OpenDPRequestModel]
146 if dummy:
File ~/work/lomas/lomas/client/lomas_client/libraries/opendp.py:62, in OpenDPClient._get_opendp_request_body(self, opendp_pipeline, fixed_delta, mechanism)
60 body_json["pipeline_type"] = OpenDpPipelineType.POLARS
61 else:
---> 62 raise InvalidQueryException(
63 f"Opendp_pipeline must either of type Measurement or LazyFrame, found {type(opendp_pipeline)}"
64 )
66 return body_json
InvalidQueryException: Opendp_pipeline must either of type Measurement or LazyFrame, found <class 'opendp.mod.Transformation'>
This is because the server will only allow measurement pipeline with differentially private results. She adds Laplacian noise to the pipeline and should be able to instantiate the pipeline.
[ ]:
var_bill_length_measurement_pipeline = (
bill_length_transformation_pipeline >>
meas.then_laplace(scale=5.0) # Noise addition mechanism instructions
)
Now that there is a measurement, she is able to apply the pipeline on the dummy dataset of the server.
[ ]:
dummy_var_res = client.opendp.query(
opendp_pipeline = var_bill_length_measurement_pipeline,
dummy=True
)
print(f"Dummy result for variance: {np.round(dummy_var_res.result.value, 2)}")
[15:28:08] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'dummy_opendp_query' with query params: {'dataset_name': 'PENGUIN', 'dummy_nb_rows': 100, 'dummy_seed': 42, 'opendp_json': '{"version": "0.12.0", "ast": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "constructor", "func": "make_chain_tt", "module": "combinators", "args": [{"_type": "constructor", "func": "make_select_column", "module": "transformations", "kwargs": {"key": "bill_length_mm", "TOA": "String"}}, {"_type": "constructor", "func": "make_split_dataframe", "module": "transformations", "kwargs": {"separator": ",", "col_names": {"_type": "list", "_items": ["species", "island", "bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g", "sex"]}}}]}, "rhs": {"_type": "constructor", "func": "then_cast_default", "module": "transformations", "kwargs": {"TOA": "f64"}}}, "rhs": {"_type": "constructor", "func": "then_clamp", "module": "transformations", "kwargs": {"bounds": [30.0, 65.0]}}}, "rhs": {"_type": "constructor", "func": "then_resize", "module": "transformations", "kwargs": {"size": 344, "constant": 44.29}}}, "rhs": {"_type": "constructor", "func": "then_variance", "module": "transformations"}}, "rhs": {"_type": "constructor", "func": "then_laplace", "module": "measurements", "kwargs": {"scale": 5.0}}}}', 'fixed_delta': None, 'pipeline_type': <OpenDpPipelineType.LEGACY: 'legacy'>, 'mechanism': <OpenDpMechanism.LAPLACE: 'laplace'>}.
Dummy result for variance: 28.69
Estimate cost with opendp
With opendp, the function opendp.cost is particularly useful to estimate the used epsilon and delta based on the scale value.
[ ]:
cost_res = client.opendp.cost(
opendp_pipeline = var_bill_length_measurement_pipeline
)
cost_res
[15:28:09] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'estimate_opendp_cost' with query params: {'dataset_name': 'PENGUIN', 'opendp_json': '{"version": "0.12.0", "ast": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "constructor", "func": "make_chain_tt", "module": "combinators", "args": [{"_type": "constructor", "func": "make_select_column", "module": "transformations", "kwargs": {"key": "bill_length_mm", "TOA": "String"}}, {"_type": "constructor", "func": "make_split_dataframe", "module": "transformations", "kwargs": {"separator": ",", "col_names": {"_type": "list", "_items": ["species", "island", "bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g", "sex"]}}}]}, "rhs": {"_type": "constructor", "func": "then_cast_default", "module": "transformations", "kwargs": {"TOA": "f64"}}}, "rhs": {"_type": "constructor", "func": "then_clamp", "module": "transformations", "kwargs": {"bounds": [30.0, 65.0]}}}, "rhs": {"_type": "constructor", "func": "then_resize", "module": "transformations", "kwargs": {"size": 344, "constant": 44.29}}}, "rhs": {"_type": "constructor", "func": "then_variance", "module": "transformations"}}, "rhs": {"_type": "constructor", "func": "then_laplace", "module": "measurements", "kwargs": {"scale": 5.0}}}}', 'fixed_delta': None, 'pipeline_type': <OpenDpPipelineType.LEGACY: 'legacy'>, 'mechanism': <OpenDpMechanism.LAPLACE: 'laplace'>}.
CostResponse(epsilon=0.7122093023265229, delta=0.0)
Execute pipeline on real dataset with opendp
She can now execute the query on the real dataset.
[ ]:
var_res = client.opendp.query(
opendp_pipeline = var_bill_length_measurement_pipeline,
)
[15:28:11] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'opendp_query' with query params: {'dataset_name': 'PENGUIN', 'opendp_json': '{"version": "0.12.0", "ast": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "partial_chain", "lhs": {"_type": "constructor", "func": "make_chain_tt", "module": "combinators", "args": [{"_type": "constructor", "func": "make_select_column", "module": "transformations", "kwargs": {"key": "bill_length_mm", "TOA": "String"}}, {"_type": "constructor", "func": "make_split_dataframe", "module": "transformations", "kwargs": {"separator": ",", "col_names": {"_type": "list", "_items": ["species", "island", "bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g", "sex"]}}}]}, "rhs": {"_type": "constructor", "func": "then_cast_default", "module": "transformations", "kwargs": {"TOA": "f64"}}}, "rhs": {"_type": "constructor", "func": "then_clamp", "module": "transformations", "kwargs": {"bounds": [30.0, 65.0]}}}, "rhs": {"_type": "constructor", "func": "then_resize", "module": "transformations", "kwargs": {"size": 344, "constant": 44.29}}}, "rhs": {"_type": "constructor", "func": "then_variance", "module": "transformations"}}, "rhs": {"_type": "constructor", "func": "then_laplace", "module": "measurements", "kwargs": {"scale": 5.0}}}}', 'fixed_delta': None, 'pipeline_type': <OpenDpPipelineType.LEGACY: 'legacy'>, 'mechanism': <OpenDpMechanism.LAPLACE: 'laplace'>}.
[ ]:
var_bill_length = np.round(var_res.result.value, 2)
print(f"Variance of bill length: {var_bill_length} (from opendp query).")
Variance of bill length: 34.66 (from opendp query).
Postprocessing: no additional privacy risk with DP
She can now do all the postprocessing that she wants with the returned data without adding any privacy risk.
[ ]:
# Get standard error
standard_error = np.sqrt(var_bill_length / NB_PENGUINS)
print(f"Standard error of bill length: {np.round(standard_error, 2)}.")
Standard error of bill length: 0.32.
[ ]:
# Compute the 95% confidence interval
ZSCORE = 1.96
lower_bound = np.round(avg_bill_length - ZSCORE * standard_error, 2)
upper_bound = np.round(avg_bill_length + ZSCORE * standard_error, 2)
print(f"The 95% confidence interval of the bill length of all penguins is [{lower_bound}, {upper_bound}].")
The 95% confidence interval of the bill length of all penguins is [43.67, 44.91].
c. Train a DP Machine Learning model with DiffPrivLib
[ ]:
from sklearn.pipeline import Pipeline
from diffprivlib import models
import pandas as pd
She now wants a model to predict the species of a penguin based on bill depth. Therefore, she uses a Random Forest classifier from DiffPrivLib library.
Prepare Random Forest Classifier pipeline on dummy with DiffPrivLib
[ ]:
feature_columns = ['bill_length_mm', 'bill_depth_mm', 'flipper_length_mm', 'body_mass_g']
target_columns = ['species']
[ ]:
def get_bounds(cols_metadata, columns):
lower = [cols_metadata[col]["lower"] for col in columns]
upper = [cols_metadata[col]["upper"] for col in columns]
return (lower, upper)
[ ]:
bounds = get_bounds(penguin_metadata['columns'], columns=feature_columns)
bounds
([30.0, 13.0, 150.0, 2000.0], [65.0, 23.0, 250.0, 7000.0])
[ ]:
dpl_pipeline = Pipeline([
('rf', models.RandomForestClassifier(
n_estimators=10,
epsilon = 2.0,
bounds=bounds,
classes=['Adelie', 'Chinstrap', 'Gentoo'])
),
])
[ ]:
dummy_response = client.diffprivlib.query(
pipeline = dpl_pipeline,
feature_columns = feature_columns,
target_columns = target_columns,
test_size = 0.2,
test_train_split_seed = 1,
dummy = True
)
model = dummy_response.result.model
model
[15:28:12] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'dummy_diffprivlib_query' with query params: {'dataset_name': 'PENGUIN', 'dummy_nb_rows': 100, 'dummy_seed': 42, 'diffprivlib_json': '{"module": "diffprivlib", "version": "0.6.6", "pipeline": [{"type": "_dpl_type:RandomForestClassifier", "name": "rf", "params": {"n_estimators": 10, "n_jobs": 1, "random_state": null, "verbose": 0, "warm_start": false, "max_depth": 5, "epsilon": 2.0, "bounds": {"_tuple": true, "_items": [[30.0, 13.0, 150.0, 2000.0], [65.0, 23.0, 250.0, 7000.0]]}, "classes": ["Adelie", "Chinstrap", "Gentoo"], "shuffle": false, "accountant": "_dpl_instance:BudgetAccountant"}}]}', 'feature_columns': ['bill_length_mm', 'bill_depth_mm', 'flipper_length_mm', 'body_mass_g'], 'target_columns': ['species'], 'test_size': 0.2, 'test_train_split_seed': 1, 'imputer_strategy': 'drop'}.
Pipeline(steps=[('rf',
RandomForestClassifier(accountant=BudgetAccountant(spent_budget=[(2.0, 0)]),
bounds=(array([ 30., 13., 150., 2000.]),
array([ 65., 23., 250., 7000.])),
classes=['Adelie', 'Chinstrap',
'Gentoo'],
epsilon=2.0))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('rf', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| n_estimators | 10 | |
| epsilon | 2.0 | |
| bounds | (array([ 30.,... 150., 2000.]), ...) | |
| classes | ['Adelie', 'Chinstrap', ...] | |
| n_jobs | 1 | |
| verbose | 0 | |
| accountant | BudgetAccount...et=[(2.0, 0)]) | |
| random_state | None | |
| max_depth | 5 | |
| warm_start | False | |
| shuffle | False |
Estimate budget of Linear Regression with DiffPrivLib
[ ]:
cost_res = client.diffprivlib.cost(
dpl_pipeline,
feature_columns = feature_columns,
target_columns = target_columns,
test_size = 0.2,
test_train_split_seed = 1
)
cost_res
[15:28:14] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'estimate_diffprivlib_cost' with query params: {'dataset_name': 'PENGUIN', 'diffprivlib_json': '{"module": "diffprivlib", "version": "0.6.6", "pipeline": [{"type": "_dpl_type:RandomForestClassifier", "name": "rf", "params": {"n_estimators": 10, "n_jobs": 1, "random_state": null, "verbose": 0, "warm_start": false, "max_depth": 5, "epsilon": 2.0, "bounds": {"_tuple": true, "_items": [[30.0, 13.0, 150.0, 2000.0], [65.0, 23.0, 250.0, 7000.0]]}, "classes": ["Adelie", "Chinstrap", "Gentoo"], "shuffle": false, "accountant": "_dpl_instance:BudgetAccountant"}}]}', 'feature_columns': ['bill_length_mm', 'bill_depth_mm', 'flipper_length_mm', 'body_mass_g'], 'target_columns': ['species'], 'test_size': 0.2, 'test_train_split_seed': 1, 'imputer_strategy': 'drop'}.
CostResponse(epsilon=2.0, delta=0.0)
Train random forest classifier on sensitive data with DiffPrivLib
[ ]:
response = client.diffprivlib.query(
pipeline = dpl_pipeline,
feature_columns = feature_columns,
target_columns = target_columns,
test_size = 0.1,
test_train_split_seed = 1,
dummy = False
)
[15:28:15] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'diffprivlib_query' with query params: {'dataset_name': 'PENGUIN', 'diffprivlib_json': '{"module": "diffprivlib", "version": "0.6.6", "pipeline": [{"type": "_dpl_type:RandomForestClassifier", "name": "rf", "params": {"n_estimators": 10, "n_jobs": 1, "random_state": null, "verbose": 0, "warm_start": false, "max_depth": 5, "epsilon": 2.0, "bounds": {"_tuple": true, "_items": [[30.0, 13.0, 150.0, 2000.0], [65.0, 23.0, 250.0, 7000.0]]}, "classes": ["Adelie", "Chinstrap", "Gentoo"], "shuffle": false, "accountant": "_dpl_instance:BudgetAccountant"}}]}', 'feature_columns': ['bill_length_mm', 'bill_depth_mm', 'flipper_length_mm', 'body_mass_g'], 'target_columns': ['species'], 'test_size': 0.1, 'test_train_split_seed': 1, 'imputer_strategy': 'drop'}.
[ ]:
# Return the mean accuracy.
model_score = response.result.score
[ ]:
f"The model has a mean accuracy of {np.round(model_score, 2)}. It is a harsh metric because we are in a multi-label classification case."
'The model has a mean accuracy of 0.11. It is a harsh metric because we are in a multi-label classification case.'
[ ]:
model = response.result.model
model
Pipeline(steps=[('rf',
RandomForestClassifier(accountant=BudgetAccountant(spent_budget=[(2.0, 0)]),
bounds=(array([ 30., 13., 150., 2000.]),
array([ 65., 23., 250., 7000.])),
classes=['Adelie', 'Chinstrap',
'Gentoo'],
epsilon=2.0))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Parameters
| steps | [('rf', ...)] | |
| transform_input | None | |
| memory | None | |
| verbose | False |
Parameters
| n_estimators | 10 | |
| epsilon | 2.0 | |
| bounds | (array([ 30.,... 150., 2000.]), ...) | |
| classes | ['Adelie', 'Chinstrap', ...] | |
| n_jobs | 1 | |
| verbose | 0 | |
| accountant | BudgetAccount...et=[(2.0, 0)]) | |
| random_state | None | |
| max_depth | 5 | |
| warm_start | False | |
| shuffle | False |
[ ]:
x_to_predict = pd.DataFrame({
'bill_length_mm': [30.0], 'bill_depth_mm': [20.0], 'flipper_length_mm': [170.0], 'body_mass_g': [5000.0]
})
predictions = model.predict(x_to_predict)[0]
f"For these feature values, the predicted species is is {predictions}."
'For these feature values, the predicted species is is Chinstrap.'
d. Get a Synthetic Dataset with Smartnoise-Synth
Finally she gets a synthetic dataset to do the rest of her analysis. She chooses to only train on a subset on 3 columns: “island”, “bill_length_mm” and “bill_depth_mm” but if we wanted she could train on the whole dataset. She also decides to use the patectgan synthesizer and keep all other default parameters.
Train patectgan synthesizer on dummy data with Smartnoise-Synth
[ ]:
res_dummy = client.smartnoise_synth.query(
synth_name="patectgan",
select_cols = ["island", "bill_length_mm", "bill_depth_mm"],
epsilon=1.0,
dummy=True,
)
dummy_synth_df = res_dummy.result.df_samples
[15:28:17] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'dummy_smartnoise_synth_query' with query params: {'dataset_name': 'PENGUIN', 'dummy_nb_rows': 100, 'dummy_seed': 42, 'synth_name': <SSynthGanSynthesizer.PATE_CTGAN: 'patectgan'>, 'epsilon': 1.0, 'delta': None, 'select_cols': ['island', 'bill_length_mm', 'bill_depth_mm'], 'synth_params': {}, 'nullable': True, 'constraints': '', 'return_model': False, 'condition': '', 'nb_samples': 200}.
[ ]:
dummy_synth_df.head()
| island | bill_length_mm | bill_depth_mm | |
|---|---|---|---|
| 0 | Dream | 47.298416 | 14.550116 |
| 1 | Dream | 57.494347 | 17.279316 |
| 2 | Dream | 45.408554 | 17.671383 |
| 3 | Biscoe | 40.397251 | 14.981976 |
| 4 | Biscoe | 50.294060 | 19.660374 |
Estimate cost of training patectgan synthesizer with Smartnoise-Synth
[ ]:
res_cost = client.smartnoise_synth.cost(
synth_name="patectgan",
epsilon=1.0,
select_cols = ["island", "bill_length_mm", "bill_depth_mm"],
)
res_cost
[15:28:20] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'estimate_smartnoise_synth_cost' with query params: {'dataset_name': 'PENGUIN', 'synth_name': <SSynthGanSynthesizer.PATE_CTGAN: 'patectgan'>, 'epsilon': 1.0, 'delta': None, 'select_cols': ['island', 'bill_length_mm', 'bill_depth_mm'], 'synth_params': {}, 'nullable': True, 'constraints': ''}.
CostResponse(epsilon=1.0, delta=0.00015673368198174188)
Train patectgan synthesizer on private data with Smartnoise-Synth
[ ]:
res = client.smartnoise_synth.query(
synth_name="patectgan",
select_cols = ["island", "bill_length_mm", "bill_depth_mm"],
epsilon=1.0,
dummy=False,
)
synth_df = res.result.df_samples
[15:28:22] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'smartnoise_synth_query' with query params: {'dataset_name': 'PENGUIN', 'synth_name': <SSynthGanSynthesizer.PATE_CTGAN: 'patectgan'>, 'epsilon': 1.0, 'delta': None, 'select_cols': ['island', 'bill_length_mm', 'bill_depth_mm'], 'synth_params': {}, 'nullable': True, 'constraints': '', 'return_model': False, 'condition': '', 'nb_samples': 200}.
[ ]:
synth_df.head()
| island | bill_length_mm | bill_depth_mm | |
|---|---|---|---|
| 0 | Biscoe | 47.812782 | 17.681633 |
| 1 | Biscoe | 44.320297 | 17.492710 |
| 2 | Biscoe | 40.627327 | 17.413399 |
| 3 | Biscoe | 54.623886 | 17.745356 |
| 4 | Torgersen | 40.598938 | 18.955317 |
Out of curiosity, she checks the average bill length and variance of bill length on this dataset.
[ ]:
synth_mean = np.round(synth_df["bill_length_mm"].mean(), 2)
synth_variance = np.round(synth_df["bill_length_mm"].var(), 2)
[ ]:
print(
f"The average with Smartnoise-SQL on private data was {avg_bill_length}.\n"
+ f"The average with Smartnoise-Synth on synthetic data is {synth_mean}."
)
The average with Smartnoise-SQL on private data was 44.29.
The average with Smartnoise-Synth on synthetic data is 48.04.
[ ]:
print(
f"The variance with opendp on private data was {var_bill_length}.\n"
+ f"The variance with Smartnoise-Synth on synthetic data is {synth_variance}."
)
The variance with opendp on private data was 34.66.
The variance with Smartnoise-Synth on synthetic data is 38.37.
Step 4: See archives of queries
She now wants to verify all the queries that she did on the real data. It is possible because an archive of all queries is kept in a secure database. With a function call she can see her queries, budget and associated responses.
[ ]:
previous_queries = client.get_previous_queries()
len(previous_queries)
[15:28:24] DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_previous_queries' with query params: {'dataset_name': 'PENGUIN'}.
DEBUG User 'dr.antartica@example.com') is making a request to url http_client.py:77 'http://localhost:48080/' at the endpoint 'get_dataset_metadata' with query params: {'dataset_name': 'PENGUIN'}.
4
[ ]:
# Smartnoise-SQL
avg_bill_length_query = previous_queries[0]
avg_bill_length_query
{'user_name': 'Dr.Antartica',
'dataset_name': 'PENGUIN',
'dp_library': 'smartnoise_sql',
'client_input': {'dataset_name': 'PENGUIN',
'query_str': 'SELECT AVG(bill_length_mm) AS avg_bill_length_mm FROM df',
'epsilon': 0.5,
'delta': 0.0001,
'mechanisms': {},
'postprocess': True},
'response': {'epsilon': 1.0,
'delta': 4.999999999999449e-05,
'requested_by': 'Dr.Antartica',
'result': {'type': 'smartnoise_sql',
'df': {'index': [0],
'columns': ['avg_bill_length_mm'],
'data': [[44.28515045992143]],
'index_names': [None],
'column_names': [None]}}},
'timestamp': 1770301686.8726397}
[ ]:
# OpenDP
var_bill_length_query = previous_queries[1]
var_bill_length_query
{'user_name': 'Dr.Antartica',
'dataset_name': 'PENGUIN',
'dp_library': 'opendp',
'client_input': {'dataset_name': 'PENGUIN',
'opendp_json': Measurement(
input_domain = AtomDomain(T=String),
input_metric = SymmetricDistance(),
output_measure = MaxDivergence),
'fixed_delta': None,
'pipeline_type': 'legacy',
'mechanism': 'laplace'},
'response': {'epsilon': 0.7122093023265229,
'delta': 0.0,
'requested_by': 'Dr.Antartica',
'result': {'type': 'opendp', 'value': 34.66151898528732}},
'timestamp': 1770301691.567336}
[ ]:
# DiffPrivLib
reg_bill_length_query = previous_queries[2]
reg_bill_length_query
{'user_name': 'Dr.Antartica',
'dataset_name': 'PENGUIN',
'dp_library': 'diffprivlib',
'client_input': {'dataset_name': 'PENGUIN',
'diffprivlib_json': '{"module": "diffprivlib", "version": "0.6.6", "pipeline": [{"type": "_dpl_type:RandomForestClassifier", "name": "rf", "params": {"n_estimators": 10, "n_jobs": 1, "random_state": null, "verbose": 0, "warm_start": false, "max_depth": 5, "epsilon": 2.0, "bounds": {"_tuple": true, "_items": [[30.0, 13.0, 150.0, 2000.0], [65.0, 23.0, 250.0, 7000.0]]}, "classes": ["Adelie", "Chinstrap", "Gentoo"], "shuffle": false, "accountant": "_dpl_instance:BudgetAccountant"}}]}',
'feature_columns': ['bill_length_mm',
'bill_depth_mm',
'flipper_length_mm',
'body_mass_g'],
'target_columns': ['species'],
'test_size': 0.1,
'test_train_split_seed': 1,
'imputer_strategy': 'drop'},
'response': {'epsilon': 2.0,
'delta': 0.0,
'requested_by': 'Dr.Antartica',
'result': {'type': 'diffprivlib',
'score': 0.11428571428571428,
'model': Pipeline(steps=[('rf',
RandomForestClassifier(accountant=BudgetAccountant(spent_budget=[(2.0, 0)]),
bounds=(array([ 30., 13., 150., 2000.]),
array([ 65., 23., 250., 7000.])),
classes=['Adelie', 'Chinstrap',
'Gentoo'],
epsilon=2.0))])}},
'timestamp': 1770301696.724773}
[ ]:
# Smartnoise-Synth
sysynth_query = previous_queries[3]
sysynth_query
{'user_name': 'Dr.Antartica',
'dataset_name': 'PENGUIN',
'dp_library': 'smartnoise_synth',
'client_input': {'dataset_name': 'PENGUIN',
'synth_name': 'patectgan',
'epsilon': 1.0,
'delta': None,
'select_cols': ['island', 'bill_length_mm', 'bill_depth_mm'],
'synth_params': {},
'nullable': True,
'constraints': '',
'return_model': False,
'condition': '',
'nb_samples': 200},
'response': {'epsilon': 1.0,
'delta': 0.00015673368198174188,
'requested_by': 'Dr.Antartica',
'result': type \
index sn_synth_samples
columns sn_synth_samples
data sn_synth_samples
index_names sn_synth_samples
column_names sn_synth_samples
df_samples
index [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,...
columns [island, bill_length_mm, bill_depth_mm]
data [[Biscoe, 47.812782287597656, 17.6816329956054...
index_names [None]
column_names [None] },
'timestamp': 1770301704.181548}